Nucleic acid dyes are a class of compounds that specifically bind to nucleic acid molecules and are commonly used in the quantitative analysis and detection of nucleic acids. These dyes are typically aromatic compounds, often containing structures such as benzene or pyridine rings. These ring structures impart unique optical properties: they absorb ultraviolet or blue light and emit fluorescence.
The composition of nucleic acid molecules
Nucleic acid molecules, such as DNA and RNA, are composed of nucleotide units. Each nucleotide consists of three parts: a five-carbon sugar, a nitrogenous base, and a phosphate group. While DNA and RNA differ in the type of five-carbon sugar (deoxyribose vs. ribose) and the types of bases they contain (DNA: adenine, thymine, guanine, cytosine; RNA: adenine, uracil, guanine, cytosine), both contain four primary bases.
mechanism
The key mechanism by which nucleic acid dyes perform their staining function lies in: They achieve specific labeling of nucleic acids by interacting with one or more bases in nucleic acids (such as intercalation, electrostatic binding, or groove binding). The binding principle is based on three mechanisms:
1. Electrostatic effect: Positively charged dyes attract negatively charged nucleic acids
2. Hydrogen bond formation: Dye molecules establish hydrogen bonds with nucleic acid bases
3. Hydrophobic effect: Non-polar regions bond to each other
When the dye binds to nucleic acids, the fluorescence intensity increases significantly (usually 20-1000 times), which is the basis for quantitative detection.
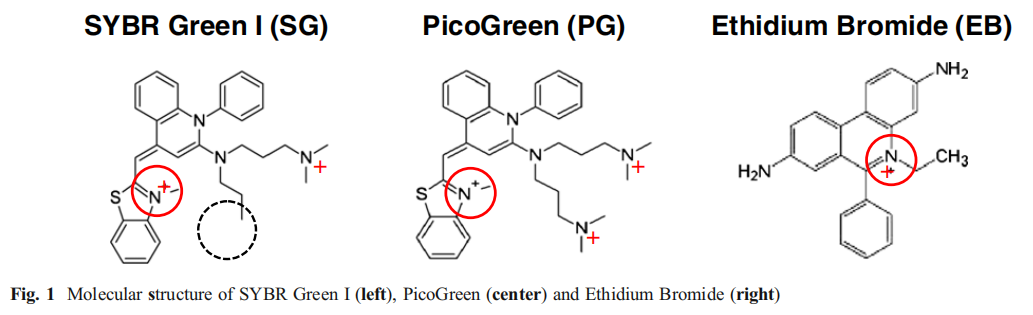
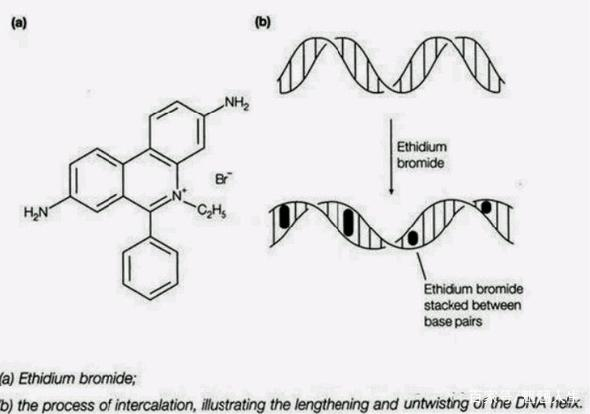
Ethidium Bromide (EB):
Features: It has a long history of application, is inexpensive, and has almost no base sequence specificity when binding to DNA. In a saturated solution with high ionic strength, approximately one EB molecule is inserted for every 2.5 bases.
Function and detection: While its intrinsic fluorescence is weak, it efficiently binds to single-stranded and double-stranded DNA. The resulting complex emits bright orange-red fluorescence under ~300nm UV excitation, increasing fluorescence intensity by 20-30 times, enabling sensitive detection of DNA bands as low as 10ng.
shortcoming: It has significant toxicity and the ability to induce DNA mutations (carcinogenicity).
status quo: It has now been replaced by a variety of non-toxic/low-toxic nucleic acid dyes.
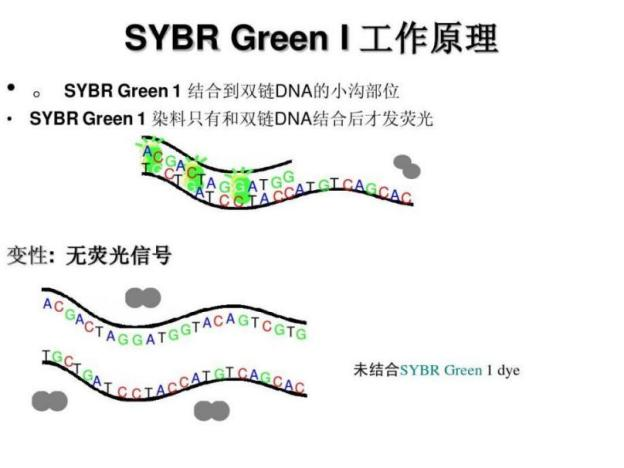
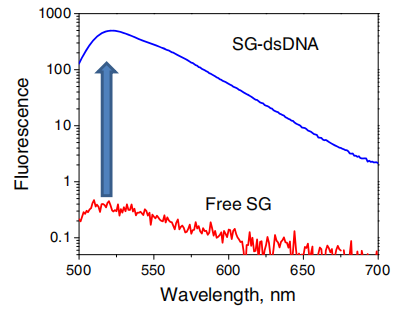
SYBR Green I:
Function and detection: The fluorescence is weak in the free state, but significantly increases upon binding to dsDNA. The fluorescence signal intensity is proportional to the amount of dsDNA and is commonly used to measure product quantity in real-time quantitative PCR (qPCR).
Optical properties: The maximum absorption wavelength is about 497nm, and the maximum emission wavelength is about 520nm (green fluorescence).
SYBR Green II:
Features: It primarily binds to RNA and single-stranded DNA (ssDNA) and is a highly sensitive RNA/ssDNA detection dye.
Function and detection: Although not RNA-specific, the fluorescence quantum yield of RNA binding (~0.54) is higher than that of DNA binding (~0.36). The fluorescence of the dye-RNA complex is not quenched by denaturants such as urea or formaldehyde, eliminating the need for denaturant washout prior to staining. It can detect as little as 100 pg of ssDNA or 2 ng of RNA bands in gels.
application: Especially suitable for SSCP analysis and RNA analysis.
SYBR Gold:
Features: It is the most sensitive dye in the SYBR series.
Function and detection: The fluorescence is enhanced more than 1000-fold after binding to nucleic acids.
applicability: It can be used to detect double-stranded DNA, single-stranded DNA and RNA.
application: Suitable for agarose gel electrophoresis, denaturing gradient gel electrophoresis (DGGE), single-strand conformational polymorphism (SSCP) studies, and other routine analyses.
SYBR Safe:
Features: It has excellent safety performance and its mutagenicity is much lower than EB.
Function and detection: The detection sensitivity is comparable to that of EB. The excitation peaks are at ~280nm and ~502nm, and the emission peak is approximately 530nm.
Advantages: After staining, the sample can be observed using a standard UV transilluminator, a visible light transilluminator, or a laser scanner. A key advantage is its compatibility with blue light transilluminators: using a blue light transilluminator avoids UV damage to DNA samples and eliminates the risk of UV damage to the human eye.
GelRed / GelGreen:
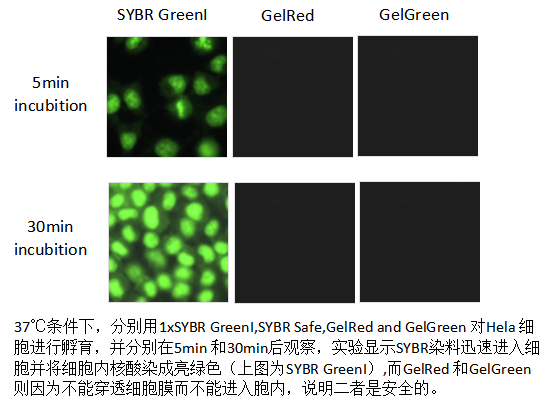
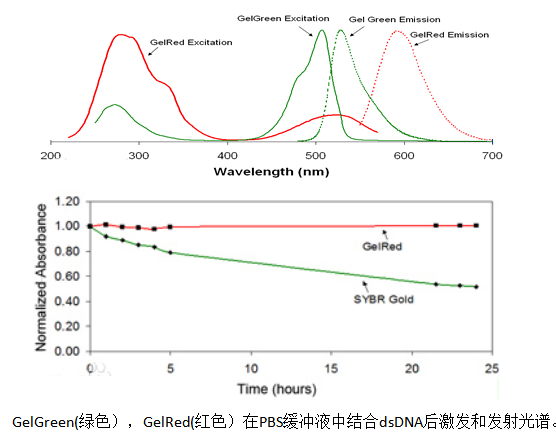
Features: GelRed and GelGreen are another important class of EB alternative dyes, with detection sensitivity no less than EB and no need for decolorization steps.
Security: Its toxicity and mutagenicity are much lower than EB, mainly due to its special chemical structure that makes it difficult to penetrate the cell membrane, thereby significantly reducing cytotoxicity.
Select suggestion:
When using a UV imager, select GelRed.
Select GelGreen when using a laser imager or wishing to observe in visible light.