Constantly having problems with your experiments? Save this comprehensive PCR troubleshooting guide!
1. False negative (no amplification product)
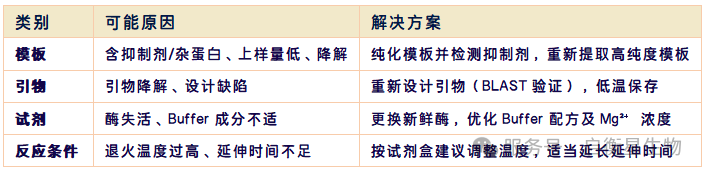
Phenomenon: There is a band in the positive control, but no band in the experimental sample
Causes and solutions:
2. False Positives
Phenomenon: Nonspecific bands appeared in the blank control
Possible causes:
The primer design is inappropriate and has homology with the target sequence
Reagent contamination: water, pipettes, etc. are contaminated with nucleic acids
Cross contamination between samples
Pollution prevention and control measures:
Environmental disinfection: high-pressure sterilization of laboratory benches and pipettes, use sterile water
Consumables management: centrifuge tubes/pipette tips are disposable, reagents (except enzymes) are sterilized by high pressure
Operation specification: Zoning operation (reagent preparation area, sample processing area, amplification area)
Technical optimization: using nested PCR or high-specificity kits
3. Tailing and diffuse banding
Phenomenon: The electrophoresis pattern is smeared or tailed
Optimization strategy:
Enzyme dosage: Reduce the amount of Taq enzyme or use hot start enzyme instead
Template quality: Purify DNA/RNA templates and remove protein/polysaccharide impurities
System balance: reduce dNTP concentration (<0.2mM), optimize Mg²⁺ concentration (1.5-2.5mM)
Number of cycles: Reduce the number of cycles (usually 25-35 times)
4. Dimers and non-specific products
Generally, bands smaller than 100 bp can be basically judged as primer dimers.
Solution:
Primer design: avoid complementarity between primers and avoid 3 consecutive G/C at the 3' end
Reaction conditions: reduce Mg²⁺ concentration and increase annealing temperature
Template amount: Increase the template input (50-100ng)
5. Core technologies for improving PCR specificity
Golden rules for primer design
Conserved region positioning: Prioritize the conserved region of the template cDNA
Length and GC content: 15-30bp, GC% <60%, no more than three consecutive G or C at the 3' end
Secondary structure: Avoid primers from forming hairpin structures or dimers
BLAST validation: Ensure that the primers have no homology to other regions of the genome
Experimental tips
Three check principles:
Check template purity (inhibitors/degradation)
Secondary primer design (BLAST/dimer)
Three-adjustment reaction system (Mg²⁺/enzyme amount/temperature)
Two must-dos:
Negative controls must be set up! Consumables must be strictly sterilized!